allofus: an R package to facilitate use of the All of Us Researcher Workbench
The Roux Institute at Northeastern University
Bouve College of Health Sciences, Northeastern University
The Roux Institute at Northeastern University
Who we are

Assistant professor
- causal inference, missing data and selection bias, pregnancy and reproductive health

Research data analyst
- health services research, neurorehabilitation, open-source software development
mentoring other researchers using All of Us data
Challenges
- Lack of programming skills
- Health researchers often use R or SAS, some use python, few use SQL
tidyverseis (the most?) popular framework for R
- All of Us data is complex
- the OMOP CDM has a steep learning curve, plus the complexity of the other data (e.g., surveys)
- many researchers are trained on cleaner, more straightforward data
- Large scale observational health research is hard
- defining cohorts with appropriate inclusion/exclusion criteria, exposure/outcome phenotypes, time under observation, etc.
- huge potential for confounding, selection bias, measurement error…
- reproducibility is critical
allofus R package
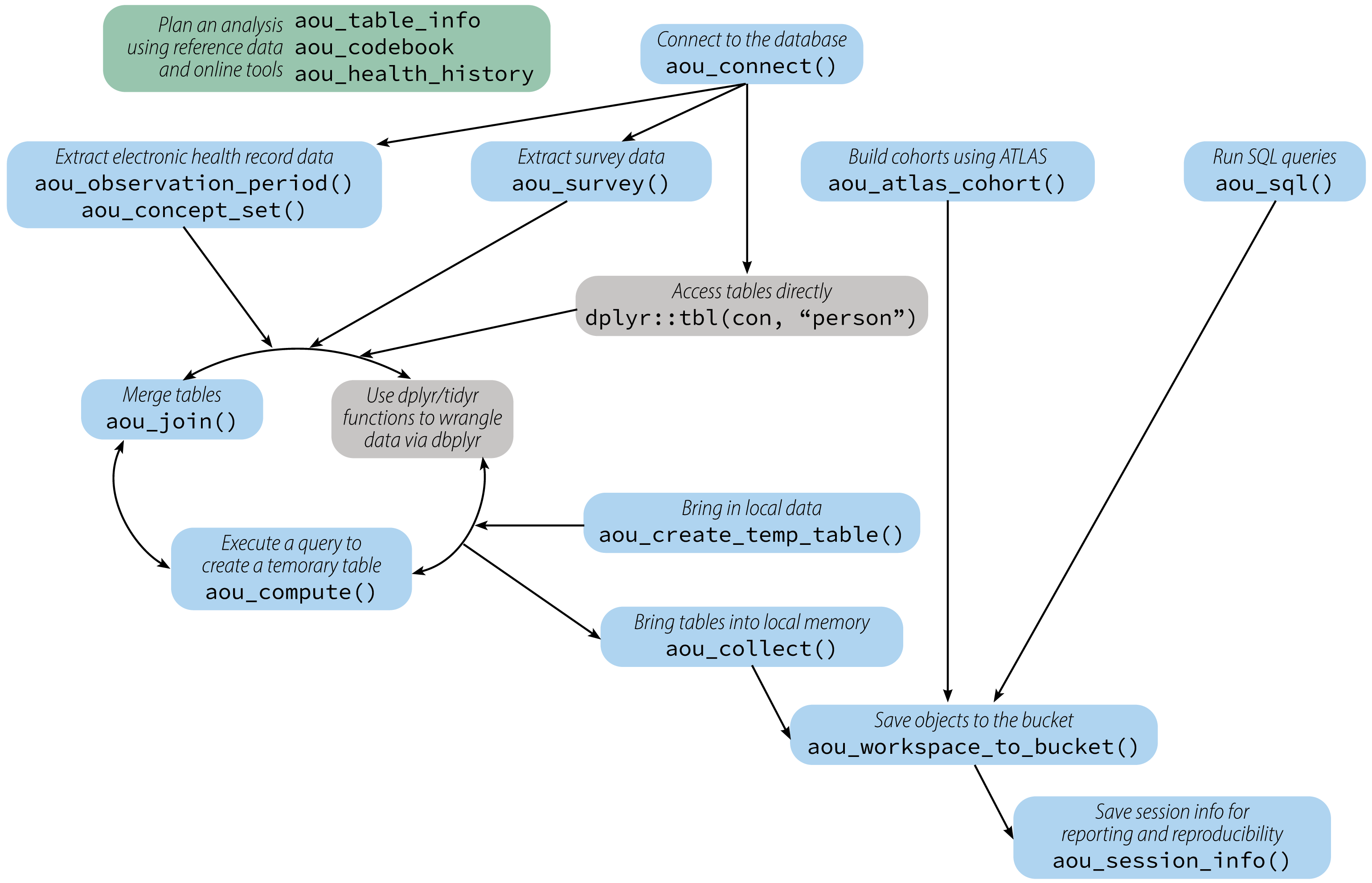
- Build on existing tools/skills
- Enable complex research
- Efficient
- Reproducible
When to start antihypertensive medications during pregnancy?
| Eligibility criteria | Pregnant, with chronic hypertension diagnosed prior to pregnancy but no prior use of antihypertensive medications |
| Baseline (time zero) | First prenatal visit |
| Treatment strategies | Initiate antihypertensive medications 1) immediately vs. 2) delayed vs. 3) never |
| Outcome | Composite of preeclampsia with severe features, medically indicated preterm birth at <35 weeks of gestation, placental abruption, or fetal or neonatal death |
When to start antihypertensive medications during pregnancy?
1. Define the cohort
2. Extract data from different domains to create a dataset with exposure, outcome, covariates
3. Analyze the data
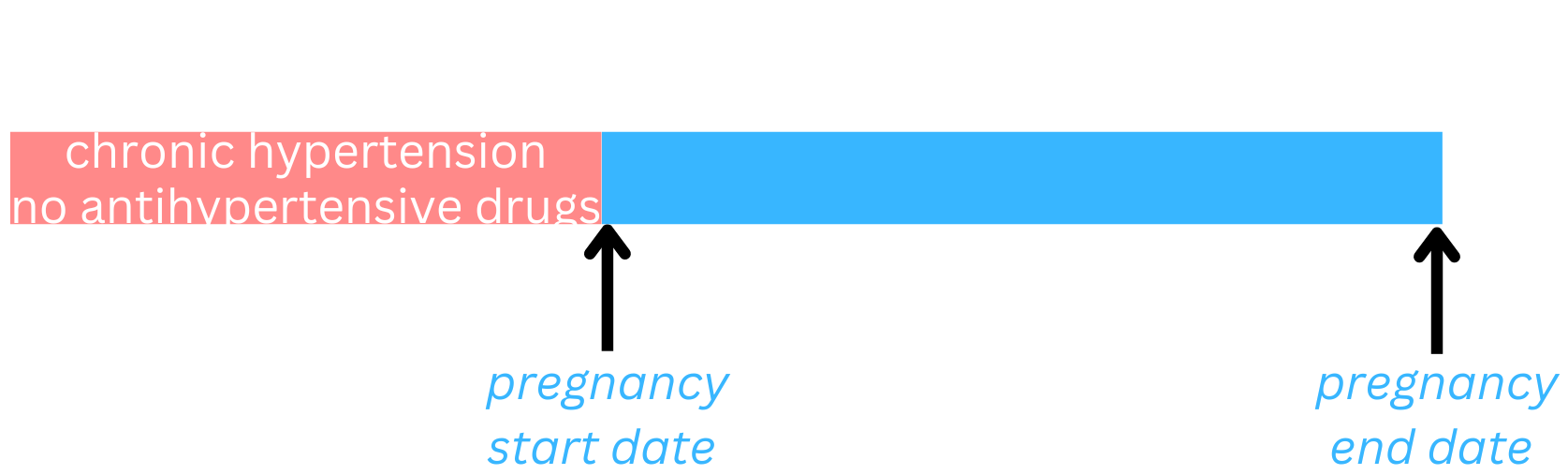
Where to begin?

Begin in R
Everything is built on a direct connection to the bigquery database
Access tables
Under the hood, dbplyr (part of the tidyverse) allows researchers to use many of the same functions on a remote table that they would use on a dataframe
[1] "person_id" "gender_concept_id"
[3] "year_of_birth" "month_of_birth"
[5] "day_of_birth" "birth_datetime"
[7] "race_concept_id" "ethnicity_concept_id"
[9] "location_id" "provider_id"
[11] "care_site_id" "person_source_value"
[13] "gender_source_value" "gender_source_concept_id"
[15] "race_source_value" "race_source_concept_id"
[17] "ethnicity_source_value" "ethnicity_source_concept_id"
[19] "state_of_residence_concept_id" "state_of_residence_source_value"
[21] "sex_at_birth_concept_id" "sex_at_birth_source_concept_id"
[23] "sex_at_birth_source_value" Quick data manipulations and computations
Users are running SQL queries without realizing they’re running SQL queries
person |>
mutate(birth_decade = cut(year_of_birth,
breaks = c(1930, 1940, 1950, 1960, 1970,
1980, 1990, 2000, 2010))) |>
count(birth_decade)# Source: SQL [9 x 2]
# Database: BigQueryConnection
birth_decade n
<chr> <int64>
1 (1960,1970] 79925
2 (1940,1950] 55220
3 (1930,1940] 13366
4 (1950,1960] 85630
5 (1970,1980] 60489
6 (1980,1990] 64452
7 (1990,2000] 50106
8 (2000,2010] 2816
9 <NA> 1453Behind the scenes
SELECT `birth_decade`, count(*) AS `n`
FROM (
SELECT
`person`.*,
CASE
WHEN (`year_of_birth` <= 1930.0) THEN NULL
WHEN (`year_of_birth` <= 1940.0) THEN '(1930,1940]'
WHEN (`year_of_birth` <= 1950.0) THEN '(1940,1950]'
WHEN (`year_of_birth` <= 1960.0) THEN '(1950,1960]'
WHEN (`year_of_birth` <= 1970.0) THEN '(1960,1970]'
WHEN (`year_of_birth` <= 1980.0) THEN '(1970,1980]'
WHEN (`year_of_birth` <= 1990.0) THEN '(1980,1990]'
WHEN (`year_of_birth` <= 2000.0) THEN '(1990,2000]'
WHEN (`year_of_birth` <= 2010.0) THEN '(2000,2010]'
WHEN (`year_of_birth` > 2010.0) THEN NULL
END AS `birth_decade`
FROM `person`
) `q01`
GROUP BY `birth_decade`Creating cohorts with Cohort Builder

Creating cohorts with allofus
reproductive_age_female <- tbl(con, "cb_search_person") |>
filter(age_at_consent >= 18 & age_at_consent <= 55,
sex_at_birth == "Female")
hypertensive_disorder <- tbl(con, "concept_ancestor") |>
filter(ancestor_concept_id == 316866) |>
inner_join(
tbl(con, "condition_occurrence"),
by = join_by(descendant_concept_id == condition_concept_id)) |>
distinct(person_id)
cohort <- reproductive_age_female |>
inner_join(hypertensive_disorder, by = join_by(person_id))
tally(cohort)# Source: SQL [1 x 1]
# Database: BigQueryConnection
n
<int64>
1 26466How does this address our challenges?
- Abstracts away all the SQL code and take advantage of existing R skills
- Enables more complex cohort definitions, including those involving non-EHR data
- Easy to carry forward the cohort to extract data and create datasets
- Readable and reproducible

Creating datasets with Cohort Builder
library(tidyverse)
library(bigrquery)
# This query represents dataset "hypertension in pregnancy" for domain "person" and was generated for All of Us Registered Tier Dataset v7
dataset_71250616_person_sql <- paste("
SELECT
person.person_id,
person.gender_concept_id,
p_gender_concept.concept_name as gender,
person.birth_datetime as date_of_birth,
person.race_concept_id,
p_race_concept.concept_name as race,
person.ethnicity_concept_id,
p_ethnicity_concept.concept_name as ethnicity,
person.sex_at_birth_concept_id,
p_sex_at_birth_concept.concept_name as sex_at_birth
FROM
`person` person
LEFT JOIN
`concept` p_gender_concept
ON person.gender_concept_id = p_gender_concept.concept_id
LEFT JOIN
`concept` p_race_concept
ON person.race_concept_id = p_race_concept.concept_id
LEFT JOIN
`concept` p_ethnicity_concept
ON person.ethnicity_concept_id = p_ethnicity_concept.concept_id
LEFT JOIN
`concept` p_sex_at_birth_concept
ON person.sex_at_birth_concept_id = p_sex_at_birth_concept.concept_id
WHERE
person.PERSON_ID IN (SELECT
distinct person_id
FROM
`cb_search_person` cb_search_person
WHERE
cb_search_person.person_id IN (SELECT
person_id
FROM
`cb_search_person` p
WHERE
age_at_consent BETWEEN 18 AND 50
AND cb_search_person.person_id IN (SELECT
person_id
FROM
`person` p
WHERE
sex_at_birth_concept_id IN (45878463) )
AND cb_search_person.person_id IN (SELECT
criteria.person_id
FROM
(SELECT
DISTINCT person_id, entry_date, concept_id
FROM
`cb_search_all_events`
WHERE
(concept_id IN(SELECT
DISTINCT c.concept_id
FROM
`cb_criteria` c
JOIN
(SELECT
CAST(cr.id as string) AS id
FROM
`cb_criteria` cr
WHERE
concept_id IN (316866)
AND full_text LIKE '%_rank1]%' ) a
ON (c.path LIKE CONCAT('%.', a.id, '.%')
OR c.path LIKE CONCAT('%.', a.id)
OR c.path LIKE CONCAT(a.id, '.%')
OR c.path = a.id)
WHERE
is_standard = 1
AND is_selectable = 1)
AND is_standard = 1 )) criteria ) )", sep="")
# Formulate a Cloud Storage destination path for the data exported from BigQuery.
# NOTE: By default data exported multiple times on the same day will overwrite older copies.
# But data exported on a different days will write to a new location so that historical
# copies can be kept as the dataset definition is changed.
person_71250616_path <- file.path(
Sys.getenv("WORKSPACE_BUCKET"),
"bq_exports",
Sys.getenv("OWNER_EMAIL"),
strftime(lubridate::now(), "%Y%m%d"), # Comment out this line if you want the export to always overwrite.
"person_71250616",
"person_71250616_*.csv")
message(str_glue('The data will be written to {person_71250616_path}. Use this path when reading ',
'the data into your notebooks in the future.'))
# Perform the query and export the dataset to Cloud Storage as CSV files.
# NOTE: You only need to run `bq_table_save` once. After that, you can
# just read data from the CSVs in Cloud Storage.
bq_table_save(
bq_dataset_query(Sys.getenv("WORKSPACE_CDR"), dataset_71250616_person_sql, billing = Sys.getenv("GOOGLE_PROJECT")),
person_71250616_path,
destination_format = "CSV")
# Read the data directly from Cloud Storage into memory.
# NOTE: Alternatively you can `gsutil -m cp {person_71250616_path}` to copy these files
# to the Jupyter disk.
read_bq_export_from_workspace_bucket <- function(export_path) {
col_types <- cols(gender = col_character(), race = col_character(), ethnicity = col_character(), sex_at_birth = col_character())
bind_rows(
map(system2('gsutil', args = c('ls', export_path), stdout = TRUE, stderr = TRUE),
function(csv) {
message(str_glue('Loading {csv}.'))
chunk <- read_csv(pipe(str_glue('gsutil cat {csv}')), col_types = col_types, show_col_types = FALSE)
if (is.null(col_types)) {
col_types <- spec(chunk)
}
chunk
}))
}
dataset_71250616_person_df <- read_bq_export_from_workspace_bucket(person_71250616_path)
dim(dataset_71250616_person_df)
head(dataset_71250616_person_df, 5)
library(tidyverse)
library(bigrquery)
# This query represents dataset "hypertension in pregnancy" for domain "condition" and was generated for All of Us Registered Tier Dataset v7
dataset_71250616_condition_sql <- paste("
SELECT
c_occurrence.person_id,
c_occurrence.condition_concept_id,
c_standard_concept.concept_name as standard_concept_name,
c_standard_concept.concept_code as standard_concept_code,
c_standard_concept.vocabulary_id as standard_vocabulary,
c_occurrence.condition_start_datetime,
c_occurrence.condition_end_datetime,
c_occurrence.condition_type_concept_id,
c_type.concept_name as condition_type_concept_name,
c_occurrence.stop_reason,
c_occurrence.visit_occurrence_id,
visit.concept_name as visit_occurrence_concept_name,
c_occurrence.condition_source_value,
c_occurrence.condition_source_concept_id,
c_source_concept.concept_name as source_concept_name,
c_source_concept.concept_code as source_concept_code,
c_source_concept.vocabulary_id as source_vocabulary,
c_occurrence.condition_status_source_value,
c_occurrence.condition_status_concept_id,
c_status.concept_name as condition_status_concept_name
FROM
( SELECT
*
FROM
`condition_occurrence` c_occurrence
WHERE
(
condition_concept_id IN (SELECT
DISTINCT c.concept_id
FROM
`cb_criteria` c
JOIN
(SELECT
CAST(cr.id as string) AS id
FROM
`cb_criteria` cr
WHERE
concept_id IN (132685, 133816, 134414, 135601, 136743, 137613, 138811, 141084, 314090, 35622939, 4034096, 4057976, 4116344, 4283352, 433536, 438490, 439077, 439393, 443700)
AND full_text LIKE '%_rank1]%' ) a
ON (c.path LIKE CONCAT('%.', a.id, '.%')
OR c.path LIKE CONCAT('%.', a.id)
OR c.path LIKE CONCAT(a.id, '.%')
OR c.path = a.id)
WHERE
is_standard = 1
AND is_selectable = 1)
)
AND (
c_occurrence.PERSON_ID IN (SELECT
distinct person_id
FROM
`cb_search_person` cb_search_person
WHERE
cb_search_person.person_id IN (SELECT
person_id
FROM
`cb_search_person` p
WHERE
age_at_consent BETWEEN 18 AND 50
AND cb_search_person.person_id IN (SELECT
person_id
FROM
`person` p
WHERE
sex_at_birth_concept_id IN (45878463) )
AND cb_search_person.person_id IN (SELECT
criteria.person_id
FROM
(SELECT
DISTINCT person_id, entry_date, concept_id
FROM
`cb_search_all_events`
WHERE
(concept_id IN(SELECT
DISTINCT c.concept_id
FROM
`cb_criteria` c
JOIN
(SELECT
CAST(cr.id as string) AS id
FROM
`cb_criteria` cr
WHERE
concept_id IN (316866)
AND full_text LIKE '%_rank1]%' ) a
ON (c.path LIKE CONCAT('%.', a.id, '.%')
OR c.path LIKE CONCAT('%.', a.id)
OR c.path LIKE CONCAT(a.id, '.%')
OR c.path = a.id)
WHERE
is_standard = 1
AND is_selectable = 1)
AND is_standard = 1 )) criteria ) )
)) c_occurrence
LEFT JOIN
`concept` c_standard_concept
ON c_occurrence.condition_concept_id = c_standard_concept.concept_id
LEFT JOIN
`concept` c_type
ON c_occurrence.condition_type_concept_id = c_type.concept_id
LEFT JOIN
`visit_occurrence` v
ON c_occurrence.visit_occurrence_id = v.visit_occurrence_id
LEFT JOIN
`concept` visit
ON v.visit_concept_id = visit.concept_id
LEFT JOIN
`concept` c_source_concept
ON c_occurrence.condition_source_concept_id = c_source_concept.concept_id
LEFT JOIN
`concept` c_status
ON c_occurrence.condition_status_concept_id = c_status.concept_id", sep="")
# Formulate a Cloud Storage destination path for the data exported from BigQuery.
# NOTE: By default data exported multiple times on the same day will overwrite older copies.
# But data exported on a different days will write to a new location so that historical
# copies can be kept as the dataset definition is changed.
condition_71250616_path <- file.path(
Sys.getenv("WORKSPACE_BUCKET"),
"bq_exports",
Sys.getenv("OWNER_EMAIL"),
strftime(lubridate::now(), "%Y%m%d"), # Comment out this line if you want the export to always overwrite.
"condition_71250616",
"condition_71250616_*.csv")
message(str_glue('The data will be written to {condition_71250616_path}. Use this path when reading ',
'the data into your notebooks in the future.'))
# Perform the query and export the dataset to Cloud Storage as CSV files.
# NOTE: You only need to run `bq_table_save` once. After that, you can
# just read data from the CSVs in Cloud Storage.
bq_table_save(
bq_dataset_query(Sys.getenv("WORKSPACE_CDR"), dataset_71250616_condition_sql, billing = Sys.getenv("GOOGLE_PROJECT")),
condition_71250616_path,
destination_format = "CSV")
# Read the data directly from Cloud Storage into memory.
# NOTE: Alternatively you can `gsutil -m cp {condition_71250616_path}` to copy these files
# to the Jupyter disk.
read_bq_export_from_workspace_bucket <- function(export_path) {
col_types <- cols(standard_concept_name = col_character(), standard_concept_code = col_character(), standard_vocabulary = col_character(), condition_type_concept_name = col_character(), stop_reason = col_character(), visit_occurrence_concept_name = col_character(), condition_source_value = col_character(), source_concept_name = col_character(), source_concept_code = col_character(), source_vocabulary = col_character(), condition_status_source_value = col_character(), condition_status_concept_name = col_character())
bind_rows(
map(system2('gsutil', args = c('ls', export_path), stdout = TRUE, stderr = TRUE),
function(csv) {
message(str_glue('Loading {csv}.'))
chunk <- read_csv(pipe(str_glue('gsutil cat {csv}')), col_types = col_types, show_col_types = FALSE)
if (is.null(col_types)) {
col_types <- spec(chunk)
}
chunk
}))
}
dataset_71250616_condition_df <- read_bq_export_from_workspace_bucket(condition_71250616_path)
dim(dataset_71250616_condition_df)
head(dataset_71250616_condition_df, 5)
library(tidyverse)
library(bigrquery)
# This query represents dataset "hypertension in pregnancy" for domain "measurement" and was generated for All of Us Registered Tier Dataset v7
dataset_71250616_measurement_sql <- paste("
SELECT
measurement.person_id,
measurement.measurement_concept_id,
m_standard_concept.concept_name as standard_concept_name,
m_standard_concept.concept_code as standard_concept_code,
m_standard_concept.vocabulary_id as standard_vocabulary,
measurement.measurement_datetime,
measurement.measurement_type_concept_id,
m_type.concept_name as measurement_type_concept_name,
measurement.operator_concept_id,
m_operator.concept_name as operator_concept_name,
measurement.value_as_number,
measurement.value_as_concept_id,
m_value.concept_name as value_as_concept_name,
measurement.unit_concept_id,
m_unit.concept_name as unit_concept_name,
measurement.range_low,
measurement.range_high,
measurement.visit_occurrence_id,
m_visit.concept_name as visit_occurrence_concept_name,
measurement.measurement_source_value,
measurement.measurement_source_concept_id,
m_source_concept.concept_name as source_concept_name,
m_source_concept.concept_code as source_concept_code,
m_source_concept.vocabulary_id as source_vocabulary,
measurement.unit_source_value,
measurement.value_source_value
FROM
( SELECT
*
FROM
`measurement` measurement
WHERE
(
measurement_concept_id IN (SELECT
DISTINCT c.concept_id
FROM
`cb_criteria` c
JOIN
(SELECT
CAST(cr.id as string) AS id
FROM
`cb_criteria` cr
WHERE
concept_id IN (21490851, 21490853, 3004249, 3005606, 3009395, 3012526, 3012888, 3013940, 3017490, 3018586, 3018592, 3018822, 3019962, 3027598, 3028737, 3031203, 3034703, 3035856, 36716965, 4060834, 40758413, 4152194, 4154790, 4232915, 4239021, 4248524, 4298393, 4302410, 44789315, 44789316)
AND full_text LIKE '%_rank1]%' ) a
ON (c.path LIKE CONCAT('%.', a.id, '.%')
OR c.path LIKE CONCAT('%.', a.id)
OR c.path LIKE CONCAT(a.id, '.%')
OR c.path = a.id)
WHERE
is_standard = 1
AND is_selectable = 1)
)
AND (
measurement.PERSON_ID IN (SELECT
distinct person_id
FROM
`cb_search_person` cb_search_person
WHERE
cb_search_person.person_id IN (SELECT
person_id
FROM
`cb_search_person` p
WHERE
age_at_consent BETWEEN 18 AND 50
AND cb_search_person.person_id IN (SELECT
person_id
FROM
`person` p
WHERE
sex_at_birth_concept_id IN (45878463) )
AND cb_search_person.person_id IN (SELECT
criteria.person_id
FROM
(SELECT
DISTINCT person_id, entry_date, concept_id
FROM
`cb_search_all_events`
WHERE
(concept_id IN(SELECT
DISTINCT c.concept_id
FROM
`cb_criteria` c
JOIN
(SELECT
CAST(cr.id as string) AS id
FROM
`cb_criteria` cr
WHERE
concept_id IN (316866)
AND full_text LIKE '%_rank1]%' ) a
ON (c.path LIKE CONCAT('%.', a.id, '.%')
OR c.path LIKE CONCAT('%.', a.id)
OR c.path LIKE CONCAT(a.id, '.%')
OR c.path = a.id)
WHERE
is_standard = 1
AND is_selectable = 1)
AND is_standard = 1 )) criteria ) )
)) measurement
LEFT JOIN
`concept` m_standard_concept
ON measurement.measurement_concept_id = m_standard_concept.concept_id
LEFT JOIN
`concept` m_type
ON measurement.measurement_type_concept_id = m_type.concept_id
LEFT JOIN
`concept` m_operator
ON measurement.operator_concept_id = m_operator.concept_id
LEFT JOIN
`concept` m_value
ON measurement.value_as_concept_id = m_value.concept_id
LEFT JOIN
`concept` m_unit
ON measurement.unit_concept_id = m_unit.concept_id
LEFT JOIn
`visit_occurrence` v
ON measurement.visit_occurrence_id = v.visit_occurrence_id
LEFT JOIN
`concept` m_visit
ON v.visit_concept_id = m_visit.concept_id
LEFT JOIN
`concept` m_source_concept
ON measurement.measurement_source_concept_id = m_source_concept.concept_id", sep="")
# Formulate a Cloud Storage destination path for the data exported from BigQuery.
# NOTE: By default data exported multiple times on the same day will overwrite older copies.
# But data exported on a different days will write to a new location so that historical
# copies can be kept as the dataset definition is changed.
measurement_71250616_path <- file.path(
Sys.getenv("WORKSPACE_BUCKET"),
"bq_exports",
Sys.getenv("OWNER_EMAIL"),
strftime(lubridate::now(), "%Y%m%d"), # Comment out this line if you want the export to always overwrite.
"measurement_71250616",
"measurement_71250616_*.csv")
message(str_glue('The data will be written to {measurement_71250616_path}. Use this path when reading ',
'the data into your notebooks in the future.'))
# Perform the query and export the dataset to Cloud Storage as CSV files.
# NOTE: You only need to run `bq_table_save` once. After that, you can
# just read data from the CSVs in Cloud Storage.
bq_table_save(
bq_dataset_query(Sys.getenv("WORKSPACE_CDR"), dataset_71250616_measurement_sql, billing = Sys.getenv("GOOGLE_PROJECT")),
measurement_71250616_path,
destination_format = "CSV")
# Read the data directly from Cloud Storage into memory.
# NOTE: Alternatively you can `gsutil -m cp {measurement_71250616_path}` to copy these files
# to the Jupyter disk.
read_bq_export_from_workspace_bucket <- function(export_path) {
col_types <- cols(standard_concept_name = col_character(), standard_concept_code = col_character(), standard_vocabulary = col_character(), measurement_type_concept_name = col_character(), operator_concept_name = col_character(), value_as_concept_name = col_character(), unit_concept_name = col_character(), visit_occurrence_concept_name = col_character(), measurement_source_value = col_character(), source_concept_name = col_character(), source_concept_code = col_character(), source_vocabulary = col_character(), unit_source_value = col_character(), value_source_value = col_character())
bind_rows(
map(system2('gsutil', args = c('ls', export_path), stdout = TRUE, stderr = TRUE),
function(csv) {
message(str_glue('Loading {csv}.'))
chunk <- read_csv(pipe(str_glue('gsutil cat {csv}')), col_types = col_types, show_col_types = FALSE)
if (is.null(col_types)) {
col_types <- spec(chunk)
}
chunk
}))
}
dataset_71250616_measurement_df <- read_bq_export_from_workspace_bucket(measurement_71250616_path)
dim(dataset_71250616_measurement_df)
head(dataset_71250616_measurement_df, 5)
library(tidyverse)
library(bigrquery)
# This query represents dataset "hypertension in pregnancy" for domain "drug" and was generated for All of Us Registered Tier Dataset v7
dataset_71250616_drug_sql <- paste("
SELECT
d_exposure.person_id,
d_exposure.drug_concept_id,
d_standard_concept.concept_name as standard_concept_name,
d_standard_concept.concept_code as standard_concept_code,
d_standard_concept.vocabulary_id as standard_vocabulary,
d_exposure.drug_exposure_start_datetime,
d_exposure.drug_exposure_end_datetime,
d_exposure.verbatim_end_date,
d_exposure.drug_type_concept_id,
d_type.concept_name as drug_type_concept_name,
d_exposure.stop_reason,
d_exposure.refills,
d_exposure.quantity,
d_exposure.days_supply,
d_exposure.sig,
d_exposure.route_concept_id,
d_route.concept_name as route_concept_name,
d_exposure.lot_number,
d_exposure.visit_occurrence_id,
d_visit.concept_name as visit_occurrence_concept_name,
d_exposure.drug_source_value,
d_exposure.drug_source_concept_id,
d_source_concept.concept_name as source_concept_name,
d_source_concept.concept_code as source_concept_code,
d_source_concept.vocabulary_id as source_vocabulary,
d_exposure.route_source_value,
d_exposure.dose_unit_source_value
FROM
( SELECT
*
FROM
`drug_exposure` d_exposure
WHERE
(
drug_concept_id IN (SELECT
DISTINCT ca.descendant_id
FROM
`cb_criteria_ancestor` ca
JOIN
(SELECT
DISTINCT c.concept_id
FROM
`cb_criteria` c
JOIN
(SELECT
CAST(cr.id as string) AS id
FROM
`cb_criteria` cr
WHERE
concept_id IN (21601664, 21601744)
AND full_text LIKE '%_rank1]%' ) a
ON (c.path LIKE CONCAT('%.', a.id, '.%')
OR c.path LIKE CONCAT('%.', a.id)
OR c.path LIKE CONCAT(a.id, '.%')
OR c.path = a.id)
WHERE
is_standard = 1
AND is_selectable = 1) b
ON (ca.ancestor_id = b.concept_id)))
AND (d_exposure.PERSON_ID IN (SELECT
distinct person_id
FROM
`cb_search_person` cb_search_person
WHERE
cb_search_person.person_id IN (SELECT
person_id
FROM
`cb_search_person` p
WHERE
age_at_consent BETWEEN 18 AND 50
AND cb_search_person.person_id IN (SELECT
person_id
FROM
`person` p
WHERE
sex_at_birth_concept_id IN (45878463) )
AND cb_search_person.person_id IN (SELECT
criteria.person_id
FROM
(SELECT
DISTINCT person_id, entry_date, concept_id
FROM
`cb_search_all_events`
WHERE
(concept_id IN(SELECT
DISTINCT c.concept_id
FROM
`cb_criteria` c
JOIN
(SELECT
CAST(cr.id as string) AS id
FROM
`cb_criteria` cr
WHERE
concept_id IN (316866)
AND full_text LIKE '%_rank1]%' ) a
ON (c.path LIKE CONCAT('%.', a.id, '.%')
OR c.path LIKE CONCAT('%.', a.id)
OR c.path LIKE CONCAT(a.id, '.%')
OR c.path = a.id)
WHERE
is_standard = 1
AND is_selectable = 1)
AND is_standard = 1 )) criteria ) )
)) d_exposure
LEFT JOIN
`concept` d_standard_concept
ON d_exposure.drug_concept_id = d_standard_concept.concept_id
LEFT JOIN
`concept` d_type
ON d_exposure.drug_type_concept_id = d_type.concept_id
LEFT JOIN
`concept` d_route
ON d_exposure.route_concept_id = d_route.concept_id
LEFT JOIN
`visit_occurrence` v
ON d_exposure.visit_occurrence_id = v.visit_occurrence_id
LEFT JOIN
`concept` d_visit
ON v.visit_concept_id = d_visit.concept_id
LEFT JOIN
`concept` d_source_concept
ON d_exposure.drug_source_concept_id = d_source_concept.concept_id", sep="")
# Formulate a Cloud Storage destination path for the data exported from BigQuery.
# NOTE: By default data exported multiple times on the same day will overwrite older copies.
# But data exported on a different days will write to a new location so that historical
# copies can be kept as the dataset definition is changed.
drug_71250616_path <- file.path(
Sys.getenv("WORKSPACE_BUCKET"),
"bq_exports",
Sys.getenv("OWNER_EMAIL"),
strftime(lubridate::now(), "%Y%m%d"), # Comment out this line if you want the export to always overwrite.
"drug_71250616",
"drug_71250616_*.csv")
message(str_glue('The data will be written to {drug_71250616_path}. Use this path when reading ',
'the data into your notebooks in the future.'))
# Perform the query and export the dataset to Cloud Storage as CSV files.
# NOTE: You only need to run `bq_table_save` once. After that, you can
# just read data from the CSVs in Cloud Storage.
bq_table_save(
bq_dataset_query(Sys.getenv("WORKSPACE_CDR"), dataset_71250616_drug_sql, billing = Sys.getenv("GOOGLE_PROJECT")),
drug_71250616_path,
destination_format = "CSV")
# Read the data directly from Cloud Storage into memory.
# NOTE: Alternatively you can `gsutil -m cp {drug_71250616_path}` to copy these files
# to the Jupyter disk.
read_bq_export_from_workspace_bucket <- function(export_path) {
col_types <- cols(standard_concept_name = col_character(), standard_concept_code = col_character(), standard_vocabulary = col_character(), drug_type_concept_name = col_character(), stop_reason = col_character(), sig = col_character(), route_concept_name = col_character(), lot_number = col_character(), visit_occurrence_concept_name = col_character(), drug_source_value = col_character(), source_concept_name = col_character(), source_concept_code = col_character(), source_vocabulary = col_character(), route_source_value = col_character(), dose_unit_source_value = col_character())
bind_rows(
map(system2('gsutil', args = c('ls', export_path), stdout = TRUE, stderr = TRUE),
function(csv) {
message(str_glue('Loading {csv}.'))
chunk <- read_csv(pipe(str_glue('gsutil cat {csv}')), col_types = col_types, show_col_types = FALSE)
if (is.null(col_types)) {
col_types <- spec(chunk)
}
chunk
}))
}
dataset_71250616_drug_df <- read_bq_export_from_workspace_bucket(drug_71250616_path)
dim(dataset_71250616_drug_df)
head(dataset_71250616_drug_df, 5)Creating datasets with Cohort Builder
I have 4 CSV files (over 6 million rows) stored in my bucket that I have to read back in to do more manipulation
gs://fc-secure-5dd899cc-249c-449a-b4b1-96abcc51898b/bq_exports/
louisahsmith@researchallofus.org/20241106/person_71250616/
person_71250616_*.csv
gs://fc-secure-5dd899cc-249c-449a-b4b1-96abcc51898b/bq_exports/
louisahsmith@researchallofus.org/20241106/condition_71250616/
condition_71250616_*.csv
gs://fc-secure-5dd899cc-249c-449a-b4b1-96abcc51898b/bq_exports/
louisahsmith@researchallofus.org/20241106/measurement_71250616/
measurement_71250616_*.csv
gs://fc-secure-5dd899cc-249c-449a-b4b1-96abcc51898b/bq_exports/
louisahsmith@researchallofus.org/20241106/drug_71250616/drug_71250616_*.csvCreating datasets with allofus
concept_ids <- c(21601664, 21601744, 21490851, 21490853, 3004249, 3005606, 3009395, 3012526, 3012888, 3013940, 3017490, 3018586, 3018592, 3018822, 3019962, 3027598, 3028737, 3031203, 3034703, 3035856, 36716965, 4060834, 40758413, 4152194, 4154790, 4232915, 4239021, 4248524, 4298393, 4302410, 44789315, 44789316, 132685, 133816, 134414, 135601, 136743, 137613, 138811, 141084, 314090, 35622939, 4034096, 4057976, 4116344, 4283352, 433536, 438490, 439077, 439393, 443700)
aou_concept_set(cohort,
concepts = concept_ids,
domains = c("drug", "measurement", "condition"),
output = "all") |>
count(concept_name, sort = TRUE)# Source: SQL [?? x 2]
# Database: BigQueryConnection
# Ordered by: desc(n)
concept_name n
<chr> <int64>
1 Systolic blood pressure 2900054
2 Diastolic blood pressure 2779353
3 Mean blood pressure 162724
4 Diastolic blood pressure--sitting 97566
5 Systolic blood pressure--sitting 96330
6 Systolic blood pressure by palpation 65209
7 Blood pressure systolic and diastolic 36353
8 Invasive Systolic blood pressure 30026
9 Blood pressure panel 26178
10 Invasive Diastolic blood pressure 23961
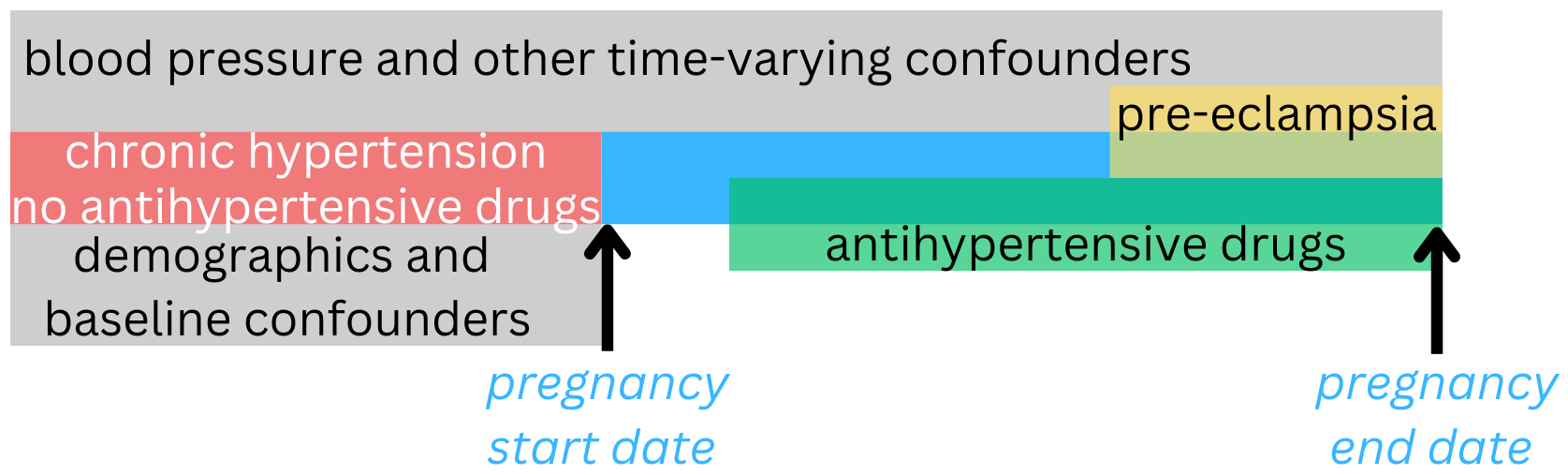
# ℹ more rowsEasily introduce temporal relationships
Let’s imagine we have a pregnancy cohort with start date! (10.1093/jamia/ocae195)
cohort_no_drugs_prior <- aou_concept_set(pregnancy_cohort,
concepts = drug_concept_ids,
start_date = NULL,
end_date = "pregnancy_start_date",
domain = "drug",
output = "indicator",
concept_set_name = "drugs_prior") |>
filter(any_drugs_prior == 0)
cohort_no_drugs_hypertension <- aou_concept_set(cohort_no_drugs_prior,
concepts = hypertension_concept_ids,
start_date = NULL,
end_date = "pregnancy_start_date",
domain = "condition",
output = "indicator",
concept_set_name = "hypertension_prior") |>
filter(hypertension_prior == 1)
Easily introduce temporal relationships
bp_during_and_prior <- aou_concept_set(cohort_no_drugs_hypertension,
concepts = bp_concept_ids,
start_date = NULL,
end_date = "pregnancy_end_date",
domain = "measurement",
output = "all")
all_drugs_during <- aou_concept_set(cohort_no_drugs_hypertension,
concepts = drug_concept_ids,
start_date = "pregnancy_start_date",
end_date = "pregnancy_end_date",
domain = "drug",
output = "all")
preeclampsia_outcomes <- aou_concept_set(cohort_no_drugs_hypertension,
concepts = preeclampsia_concept_ids,
start_date = "pregnancy_start_date",
end_date = "pregnancy_end_date",
domain = "condition",
output = "all")
Improved efficiency
- We’re extracting a lot less data because we are
- Immediately using it for what we need (eligibility criteria)
- Restricting to the time periods of interest
- The data is not actually extracted or stored until we need it in a local R session (e.g, for figures, regressions, etc.)
- Edits are straightforward and code is easily readable
vs. storing millions of rows of data in a bucket and transferring it every time you run a notebook
When you do store data, we have functions for that!
| Task | Workbench provided code snippet | allofus function |
|---|---|---|
| List files in the bucket |
|
aou_ls_bucket() |
When you do store data, we have functions for that!
| Task | Workbench provided code snippet | allofus function |
|---|---|---|
| Move a file from the bucket to workspace disk |
|
aou_bucket_to_workspace( "test.csv") |
When you do store data, we have functions for that!
| Task | Workbench provided code snippet | allofus function |
|---|---|---|
| Write a file to disk and move it to the bucket |
|
|
Integrating OHDSI software for cohort building

Survey data
When was the survey question answered?
[1] "person_id" "hypertension" "hypertension_date"Survey data
# A tibble: 6 × 2
hypertension n
<chr> <dbl>
1 <NA> 16229
2 Yes 5579
3 No 4426
4 Skip 280
5 DontKnow 49
6 PreferNotToAnswer 6- “Skip/Prefer not to answer/Don’t know” includes anyone who skipped the whole question
NArefers to participants who never saw the question.- “No” assigned to respondents who answered the question, but didn’t select “Self”
Harder to figure out appropriate denominator
# A tibble: 8 × 2
answer n
<chr> <int>
1 Including yourself, who ... (hypertension)? - Daughter 201
2 Including yourself, who ... (hypertension)? - Father 4195
3 Including yourself, who ... (hypertension)? - Grandparent 3915
4 Including yourself, who ... (hypertension)? - Mother 4480
5 Including yourself, who ... (hypertension)? - Self 5583
6 Including yourself, who ... (hypertension)? - Sibling 2384
7 Including yourself, who ... (hypertension)? - Son 213
8 PMI: Skip 1152Try to provide information to improve interpretability
ℹ One or more of the requested questions were only asked of people who responded that they had certain conditions.
→ The top-level question(s) will be added to the output to provide context about missing data as column(s) `circulatorycondition_hypertension_yes`.
Challenges (reprise)
- Lack of programming skills
- the
allofuspackage allows users to mostly avoid SQL and use the populartidyverseR framework
- the
- All of Us data is complex
- the
allofuspackage allows for simpler methods of cohort and outcomes specification using the OMOP CDM data, including survey data in All of Us
- the
- Large scale observational health research is hard
- the
allofuspackage helps try to avoid (some) mistakes and make code intent clear
- the
On the agenda
- Lack of programming skills
- Improve and extend functions and documentation
- Expand the methods in this package to python
- Ensure long-term stability and robustness for R package and python library
- All of Us data is complex
- Build a suite a specific functions for genomics, fitbit data
- Add integrations with existing OHDSI tools
- Large scale observational health research is hard
- Scale up tutorials and training materials that go beyond how to query the data
- creating causal models
- defining and validating cohorts
- understanding, identifying, and accounting for confounding and bias
- dealing with missing data
- training in appropriate statistical methods for observational health research
- Scale up tutorials and training materials that go beyond how to query the data
Thank you!
- Smith LH, Cavanaugh R. allofus: an R package to facilitate use of the All of Us Researcher Workbench. Journal of the American Medical Informatics Association. 2024;ocae198. doi: 10.1093/jamia/ocae198
- GitHub (source code, bug reports): github.com/roux-ohdsi/allofus
- Package site (tutorials, searchable codebooks): roux-ohdsi.github.io/allofus
- Email: l.smith@northeastern.edu; r.cavanaugh@northeastern.edu
